
Home
About
Contact
FAQ
Job Queue
Welcome to interact-ms !
interact-ms is a simple, webserver-based solution for you & your lab to reliably identify greater numbers of peptides from mass spec. experiments via inSPIRE, dive deeper into immunopeptidomics experiments with PEPSeek, or perform unbiased noncanonical peptide identification with PISCES.
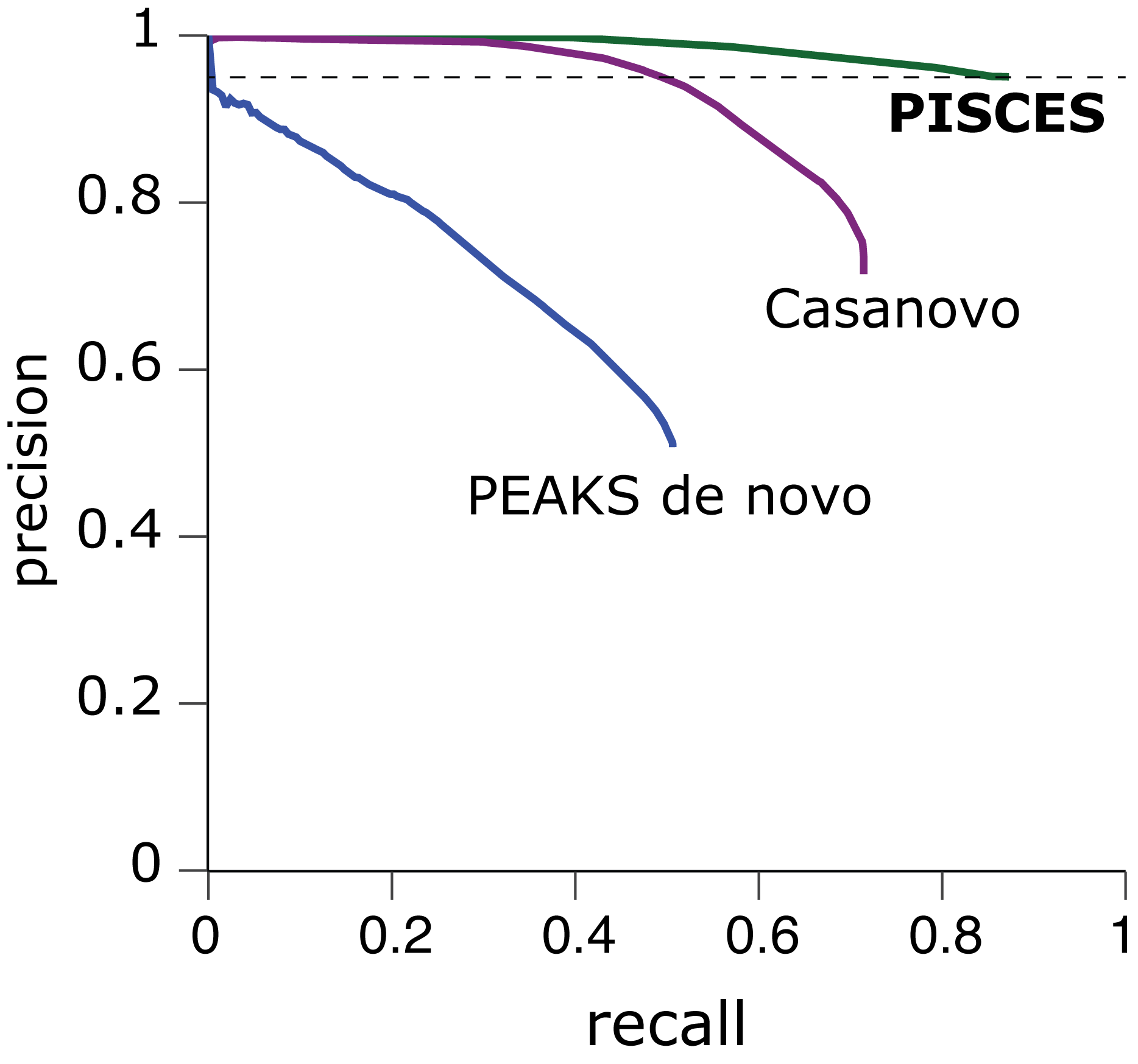
Why interact-ms?
For any mass spectrometry analysis, sensitive and reliable identification is key to maximising potential insights and discovery. interact-ms allows access to the inSPIRE rescoring pipeline for state-of-the-art identification in any experiment, and PEPSeek is a specialized tool for identification of pathogen peptides in infected immunopeptidome samples. In the latest release PISCES, a novel computational approach, eschews standard target-decoy methods and inherent biases, to provide an unbiased identification tool for noncanonical peptide identification. Further integrations can allow custom searches to be run with MSFragger, binding affinity via NetMHCpan, and quantification via Skyline to build out your analysis ecosystem entirely within interact-ms.
Our Tools
inSPIRE
PEPSeek
PISCES

Tutorials for Installation and Usage
Learn Mac/Linux installation.
Learn Windows installation.
Run PEPSeek on interact-ms.
Access & analyse PEPSeek results.